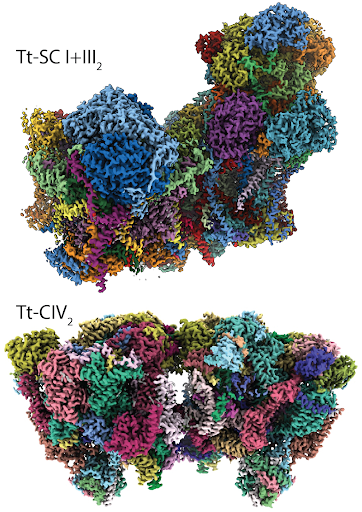
Tetrahymena thermophila is a unicellular organism that relies on a cellular respiration that is slightly different from other eukaryotes like plants and animals
By YASH RATHI — science@theaggie.org
Dr. James A. Lett is a UC Davis principal investigator in the Department of Molecular and Cellular Biology whose lab focuses on the electron transport membrane protein and structural bioenergetics. Recently, Lett’s team published research on a specific unicellular organism called Tetrahymena thermophila (Tt) to better understand the “diversity of eukaryotic core metabolism.”
Tt uses oxygen in the process of respiration, but the respiratory chain’s role is different than it is in plants, yeast and animals. Tt is commonly found swimming around ponds and uses cilia (hair-like structures) for its locomotion. It falls under a diverse group of organisms called the SARS supergroup, which has not been studied much aside from a few exceptions like malarial parasites.
Dr. Maria Maldonado, a UC Davis postdoctoral researcher at Lett’s Lab and a co-author for the study, explained how this research opened her eyes to all we do not know about respiration.
“There is a whole lot of diversity,” Maldonado said. “In Tt, we were completely shocked and surprised about the protein structures used for respiration. They are much larger [and] they exist in the form of a dimer, which is when two small units of protein combine to form one big unit. This [Tt study] is really opening our eyes about the crux of respiration. And again, this is just one organism in the tree of life; what about the whole rest of organisms?”
According to the published paper, Tt undergoes a series of chemical processes to complete its respiration with oxygen molecules used in the end of the reaction. For the most part, this is the same fundamental process of electron transport chain seen in humans and other eukaryotic organisms.
Tt has a different mechanism in the process of respiration however. In the 1970s and 1980s, researchers concluded that there were two different proteins that functioned differently in Tt. The first one is an electron-carrying protein, cytochrome C, and the second one is an oxygen-consuming enzyme at the end of the chain, terminal oxidase. Lett’s lab did research to understand why these enzymes differ in Tt, even though these two proteins were present across other studied eukaryotes.
A new approach in structural biology, called cryo-electron microscopy structural proteomics, was used by the researchers to study the Tt electron transport chain. In this approach, a large number of protein structures are worked out in a mixed sample at the same time. Cryo-electron microscopy creates a high resolution image of the proteins by freezing the given slide at an extremely low temperature. Working with mixed samples of isolated proteins from mitochondrial membranes, the team of researchers developed an algorithm to recognize different related structures in the solution of proteins.
Lett said that recent technology has sped up the process of analyzing proteins, and made it much more precise.
“New technology with respect to how we determine the protein structures has allowed us to work with much smaller samples and in new ways,” Lett said. “It has allowed us to essentially kind of explore the pathway very rapidly.”
Maldonado explained the approach the lab took to analyzing protein structures before using the algorithm.
“Key to this approach in general […] is to express one protein and then get its structure, but what we are doing is flipping it around,” Maldonado said. “We are starting with a natural sample and seeing what is there. We then purify it, put that slightly-purified sample into cryo-electron microscopy, and then try to see everything present there. This makes it much more productive using structural biology as an exploratory tool.”
Using the algorithm, they were successful in scanning hundreds of thousands of protein images and identified 277 protein structures in three large assemblies. They identified certain protein structures with specific genes that were not discovered before. These new protein structures are filling in the gaps of our previous knowledge about respiration and helping us to better understand this process.
Written by: Yash Rathi — science@theaggie.org